The Clinical Variant Ark client: pyark
Introduction¶
The Clinical Variant Ark (CVA) https://github.com/genomicsengland/clinical_variant_ark is a knowledge base for clinically relevant variants and their association to phenotypes, with fine grained detail for all the stages of interpretation, from automated variant prioritisation to manual classification. CVA holds the interpretation results of the 100K Genomes Project and in the future it will hold the results of the National Genomic Informatics System (NGIS).
The motivation of such a knowledge base are many:
- How do I compare cases from different assemblies, different pipelines, etc.?
- How do we enforce the virtuous cycle of genomic interpretation?
- How do I find cases that are similar to my case?
- How can I monitor the trends in the interpretation of cases?
The above questions are answered by providing the following features:
- Normalisation
- To feedback information both programmatically to the interpretation process and on demand through a GUI
- Maintain a set of manually curated variants and their association to phenotypes
- Implement phenotypic and genotypic similarity
- Being an analytical platform enabling cross cases comparisons and analyses
The CVA ecosystem is composed of a Java backend, backed by a MongoDB database, exposing a REST API. A user can access CVA in three ways: 1) directly through the REST API, 2) using the CVA portal from a browser or 3) using the Python client, pyark. The rest of this documentation will be focused on accessing CVA using pyark, although it may be a good documentation to understand what can be done using the REST API.
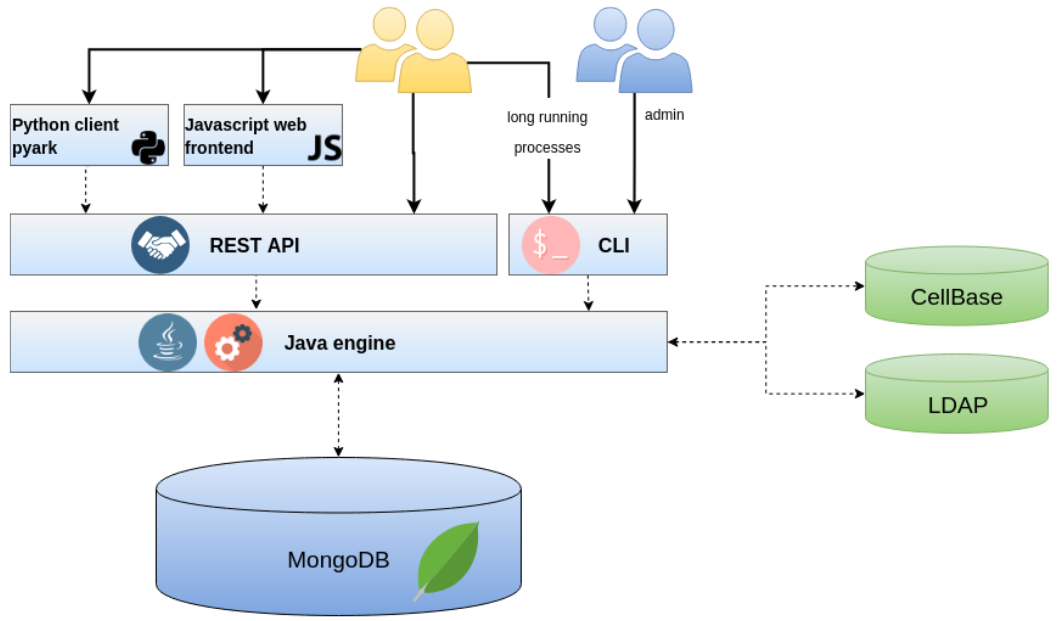
Context¶
CVA is a component in Genomics England interpretation platform. The components within this interpretation platform share a lingua franca described in the Genomics England data models, https://gelreportmodels.genomicsengland.co.uk/. This will be a useful reference manual for the data that is served by CVA.
CVA relies on three external systems:
- The interpretation API as a data provider (also known as the CIPAPI)
- Cellbase as a knowledge base of biological information, variant and gene annotations are obtained from Cellbase
- An authentication mechanism which can be either LDAP or Active Directory
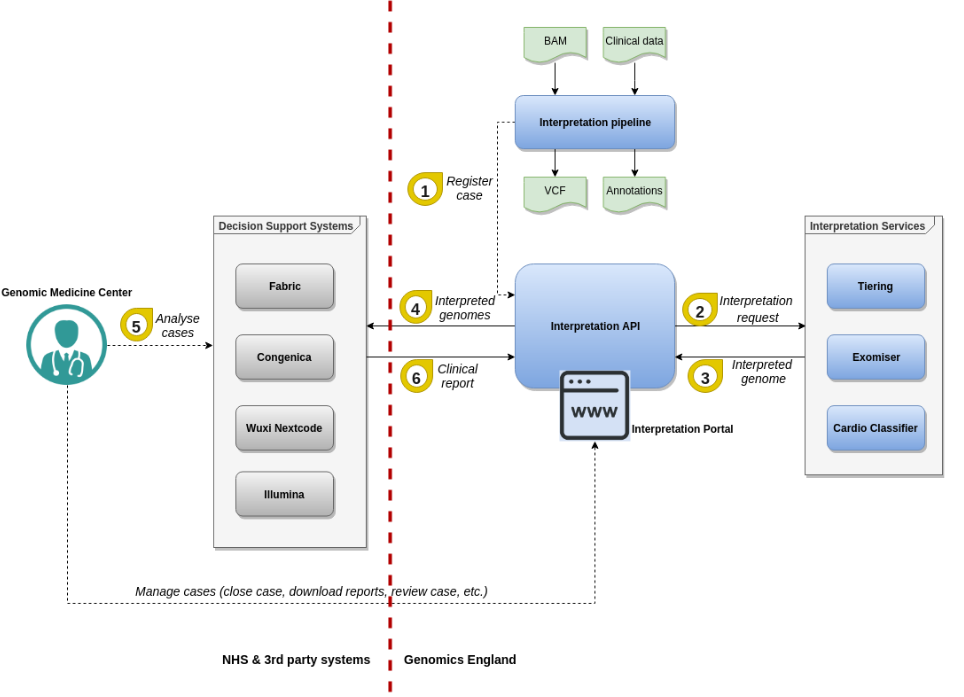
The interpretation API sends to CVA 4 pieces of data:
- The clinical data (pedigree, phenotypes, clinical indications, etc.). This data is pseudoanonymised.
- Interpreted genomes which are the result of automated variant prioritisations.
- Clinical reports which are the result of variant selection through a Decision Support System (DSS)
- Exit questionnaires which have the final classifications of variants and the final status of any given case.
The CVA client: pyark¶
Pyark is a python client to the Clinical Variant Ark (CVA) REST API. The aim of pyark is to provide easy and flexible access to the CVA dataset and enable an analytical framework. While the CVA portal aims to cover a particular use case which is helping solve cases by using existing knowledge in CVA, the aim of pyark is less precise and as such this guide will not cover everything that could be done using pyark.
For a complete guide of CVA REST API please refer to the swagger documentation available in http://your.cva.server/cva/docs.
Initialising the client¶
Fetching primary entities¶
Fetching secondary entities¶
Search and filter¶
Cohort selection¶